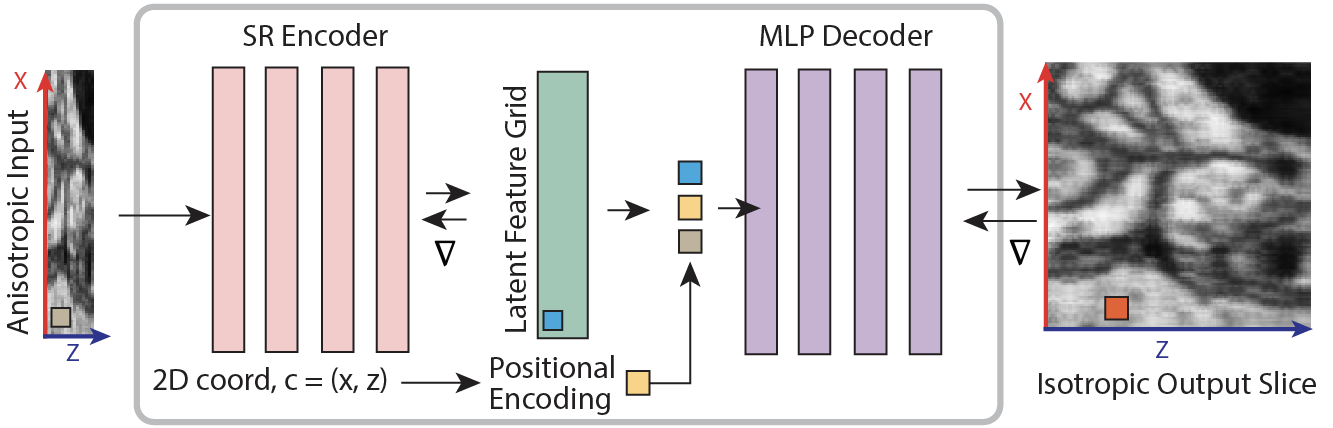
Three-dimensional (3D) microscopy often exhibit anisotropy with significantly lower resolution (up to 8x) along the z axis than along the xy axes. Computationally generating plausible isotropic resolution from anisotropic imaging data would benefit the visual analysis of large-scale volumes. This paper proposes niiv, a self-supervised method for isotropic reconstruction of 3D microscopy data that can quickly produce images at arbitrary (continuous) output resolutions. Within a neural field, the representation embeds a learned latent code that describes the implicit higher-resolution isotropic image region. Under isotropic volume assumptions, we self-supervise this representation on low-/high-resolution lateral image pairs to reconstruct an isotropic volume from low-resolution axial images. We evaluate our method on simulated and real anisotropic electron (EM) and light microscopy (LM) data. Compared to a state-of-the-art diffusion-based method, niiv shows improved reconstruction quality (+1db PSNR) and is over three orders of magnitude faster (2,000x) to infer. Specifically, niiv reconstructs a 128x128x128 voxel volume in 1/10th of a second, renderable at varying (continuous) high resolutions for display.